Name: Boguslaw Obara
Profile: Professor of Image Informatics at Newcastle University.
Email: boguslaw [dot] obara [AT] newcastle [dot] ac [dot] uk
Research and Professional Skills
Image Processing 95%About me
Boguslaw Obara is a Professor of Image Informatics in the School of Computing at Newcastle University. He is also a Turing Fellow at the Alan Turing Institute. He obtained his first master's degree in Computational Physics from Jagiellonian University, and a PhD in Image Processing from AGH University of Science and Technology.
Before joining Newcastle University as a Professor, he held research assistant positions at the Polish Academy of Sciences and at the Computer Vision Laboratory at ETH Zurich. He was a Fulbright Fellow at the Vision Research Laboratory, held postdoctoral positions at the Center for BioImage Informatics at the University of California and at the Oxford e-Research Centre at the University of Oxford, and served as an Assistant Professor, Associate Professor, and Professor in the Department of Computer Science at Durham University. Boguslaw Obara was also an AstraZeneca Visiting Professor in Image Processing and Artificial Intelligence, and Dean of Business, Innovation and Skills.
Boguslaw's research focuses on the design and implementation of advanced image analysis and processing, pattern recognition, computer vision, and machine learning solutions applied across a wide range of domains.
PhD/MRes Students:
- Core members:
- Matthew James Anderson
- Burak Kucukgoz
- Joseph Smith
- Amar Rajgor
- Ak Muhammad Rahimi Pg Hj Zahari
- Associate members:
- Ethar Alzaid
- Aaron Brooks
- Ayse Betul Cengiz
- Matthew William Chapman
- Jiacheng Cheng
- Pawel Gartner
- Hani Mohamed Osman Hasan Dirar Hasan
- Georgios Kourounis
- Karoline Leiberg
- Knectt Paulschoh Lendoye
- Yen Lau
- Ross Laws
- Qurat-ul-ain Mubarak
- George Paul
- Mahsa Vali
- Xinye Yang
- Ting Zhu
- Join us!!!
Alumni Visitors:
- 2023
- Dr Mutlu Yapici, Ankara University, Turkey.
- Dr Juan Ojeda Garcia, Spain.
- 2021
- Dr Agnieszka Stankiewicz, Poznan University of Technology, Poland.
- Srinivas Iyengar, Indian Institute of Technology, India.
- Nirmal Kumawat, Indian Institute of Technology, India.
- 2020
- Zeinab Almahdi Mohammed Haroon, African Institute for Mathematical Sciences, South Africa.
- Mayank Patel, Indian Institute of Technology, India.
- Prof. Caiyun Wang, Nanjing University of Aeronautics and Astronautics, China.
- 2019
- Romaissa Mekhzouni, ENSTA ParisTech, France.
- Yagiz Batu Saatci, Antalya Bilim University, Turkey.
- Agnieszka Stankiewicz, Poznan University of Technology, Poland.
- Cesar Alberto Cigarroa Constantino, Universidad Politecnica de Chiapas, Mexico.
- 2017
- Prof. Mark Fricker, Oxford University, UK.
- 2015
- Dr Bartosz Ziółko, AGH University of Science and Technology, Poland.
- 2014
- Prof. Stéphanie Portet, University of Manitoba, Canada.
- Agnieszka Karpińska, Cracow University of Technology, Poland.
- Dr Bartosz Schramm, Jagiellonian University, Poland.
Alumni Staff:
- Anna Bator
- Dr Pedro Cárdenas
- Dr John Brennan
- Dr Juvaria Syeda
- Dr Ning Jia
- Richard Boulderstone
- Haozheng Zhang
- Dr Nan Wang
- Dr Philip Jackson
- Dr Amar Nasrulloh
- Dr Christopher Holder
- Philip Kinghorn
- Diego Lopez
- Dr Stephen Bonner
- Jonathan Frawley
- Dr Shuaa Alharbi
- Dr Haifa AlHasson
- Dr Mahnaz Mohammadi
- Gregory Marler
- Dr Cigdem Sazak
- Brian K Isaac-Medina
- Matthew Roberts
- Dr Chris Willcocks
- Dr Ibad Kureshi
- Dr Fady Medhat
- Shwetha Nair
- Dr Geng Sun

Research
PUBLICATIONS | SOFTWARE | HARDWARE | COLLABORATION | FUNDING
Image Formation and Digitisation
Image acquisition is the first step in digital image processing and is often not properly taken into account. Our team places strong emphasis on developing a solid understanding of the image formation and digitisation processes to ensure that analyses are both reliable and meaningful, avoiding inaccurate or misleading results.
Image Processing and Pattern Recognition
Pattern recognition is used to automatically extract meaningful features from digital images. Over the past years, our team has developed a wide range of novel image informatics approaches for the recognition of blob, network, and surface patterns in multi-type, multi-scale, and multi-dimensional imaging datasets.
Best Practices for Image-Driven Solutions
Imaging data management and validated, reproducible analysis are essential to ensuring reliable, interpretable results. Our team has focused on the standardisation, integrity, and quality of image-driven information and processes—from imaging data acquisition, storage, sharing, annotation, and processing to analytics.
Research Projects
Image Processing
A Multi-Eyes Principle to Uncertainty Quantification
Inspired by the Four-Eyes principle in business and cybersecurity, we propose the Multi-Eyes principle approach, which utilises multiple deep learning models to reduce bias and potentially improve diagnostic accuracy.
Image Processing
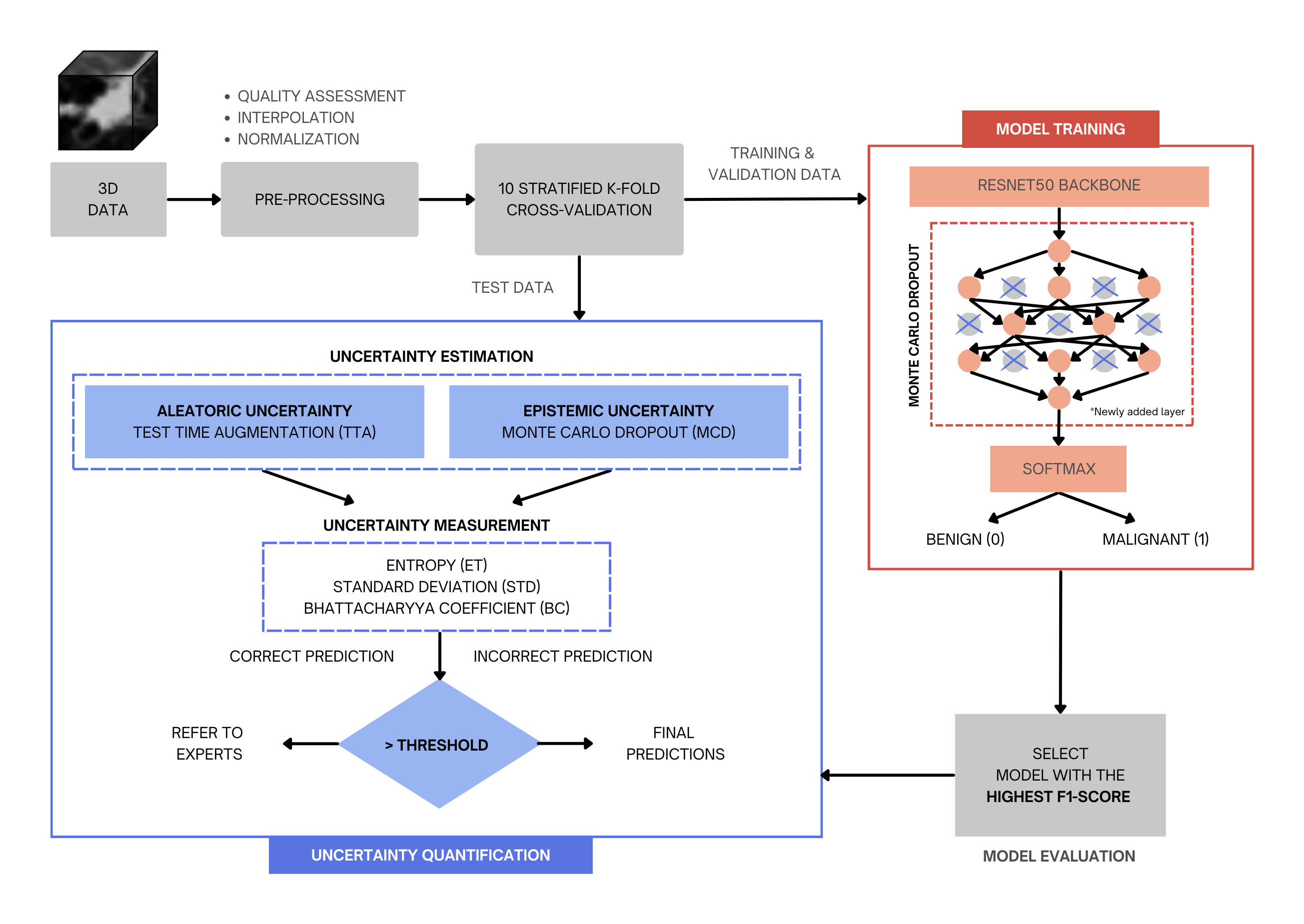
Uncertainty-aware image classification on 3D CT lung
We propose an uncertainty-aware framework that includes three phases: data preprocessing and model selection and evaluation, uncertainty quantification, and uncertainty measurement and data referral for the classification of benign and malignant nodules using 3D CT images.
Image Processing
AI Model Re-Adaptation
This framework leverages data augmentation techniques during training to enhance the generalisation of segmentation models, allowing them to adapt to newly released datasets with temporal disparity.
Image Processing
Fine-Grained Classification
An automatic training solution can find the optimal strategy for a dataset, avoiding overfitting while providing quantitative and visual explanations from the model's inputs and weights.
Image Processing
Image Quality Assessment
The project proposes a No-Reference Image Quality Assessment guided cut-off point selection strategy to enhance the performance of a fine-grained classification system. Taking the three most commonly adopted image augmentation configurations, we formulate a two-step mechanism for selecting the most discriminative subset from a given image dataset by considering both the confidence of model predictions and the density distribution of image qualities.
Image Processing
Visual Acuity Prediction
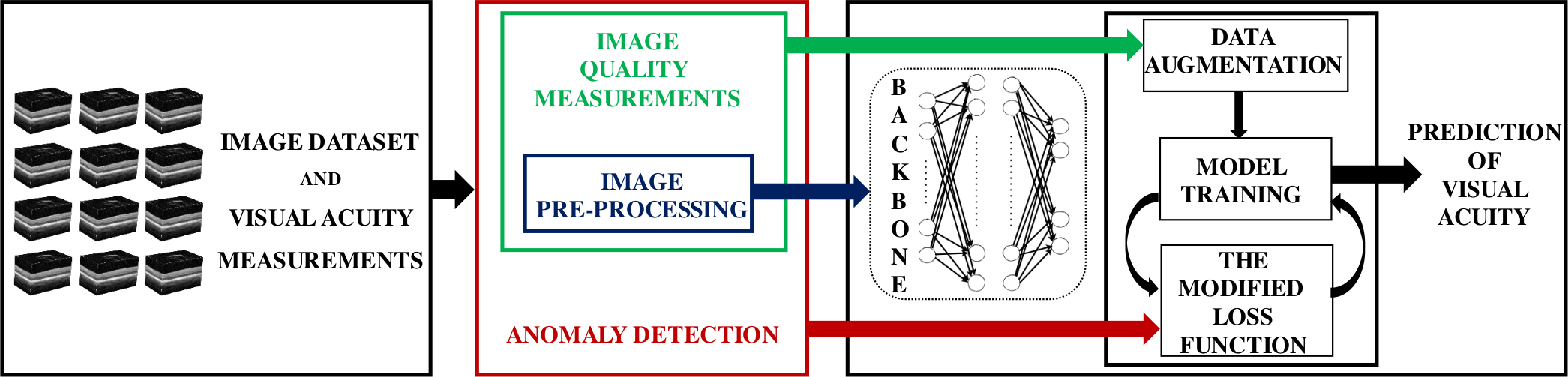
The framework is combined with a deep learning approach to automatically predict visual acuity outcomes for people undergoing surgery for idiopathic full-thickness macular holes using 3D spectral-domain optical coherence tomography images. To overcome the impact of high variation in real-world image quality on the robustness of DL models, comprehensive imaging data pre-processing, quality assurance, and anomaly detection procedures were utilised.
Image Processing
Laryngeal Cancers Biomarkers
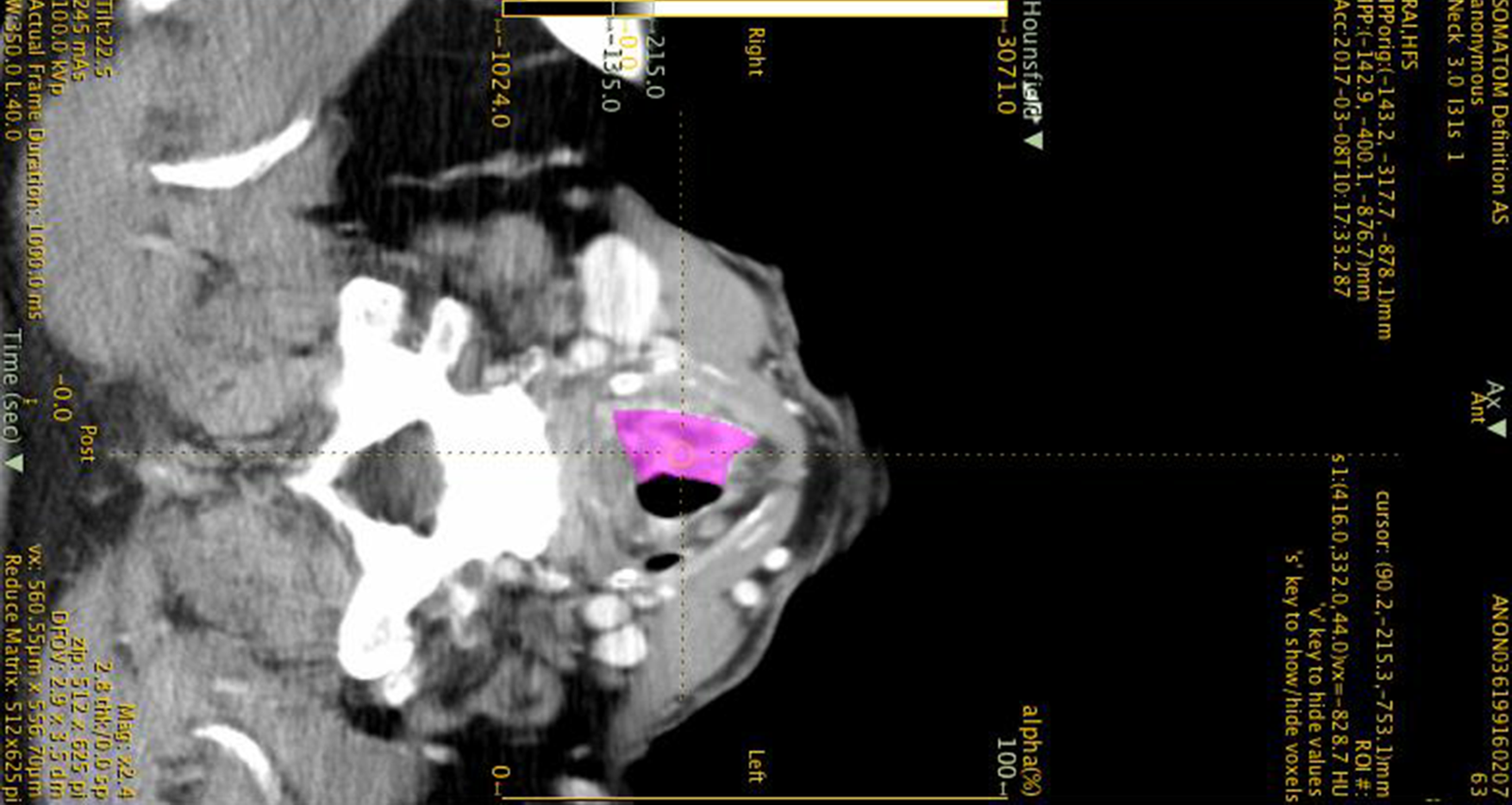
Advanced laryngeal cancers are clinically complex; there is a paucity of modern decision-making models to guide tumour-specific management. This pilot study aims to identify computed tomography-based radiomic features that may predict survival and enhance prognostication.
Image Processing
Vitreo-Retinal Surgery
The NHS became the first national health system pledging to attain 'net zero' emissions, a target it aims to achieve by 2040. Ophthalmologists have a role in making changes to mitigate our carbon footprint by considering a use of fluorinated gases ('F-gases').
Image Processing
Lung Cancer Classification
Radiologists should have complete confidence in the prediction of the model during the screening. Therefore, to address this issue, we have developed an uncertainty-aware model framework not only to classify lung cancer nodules from 3D computed tomography images, but also to estimate the associated uncertainty in the prediction of the model.
Image Processing
Biomedical Data Annotation: An OCT Imaging Case Study
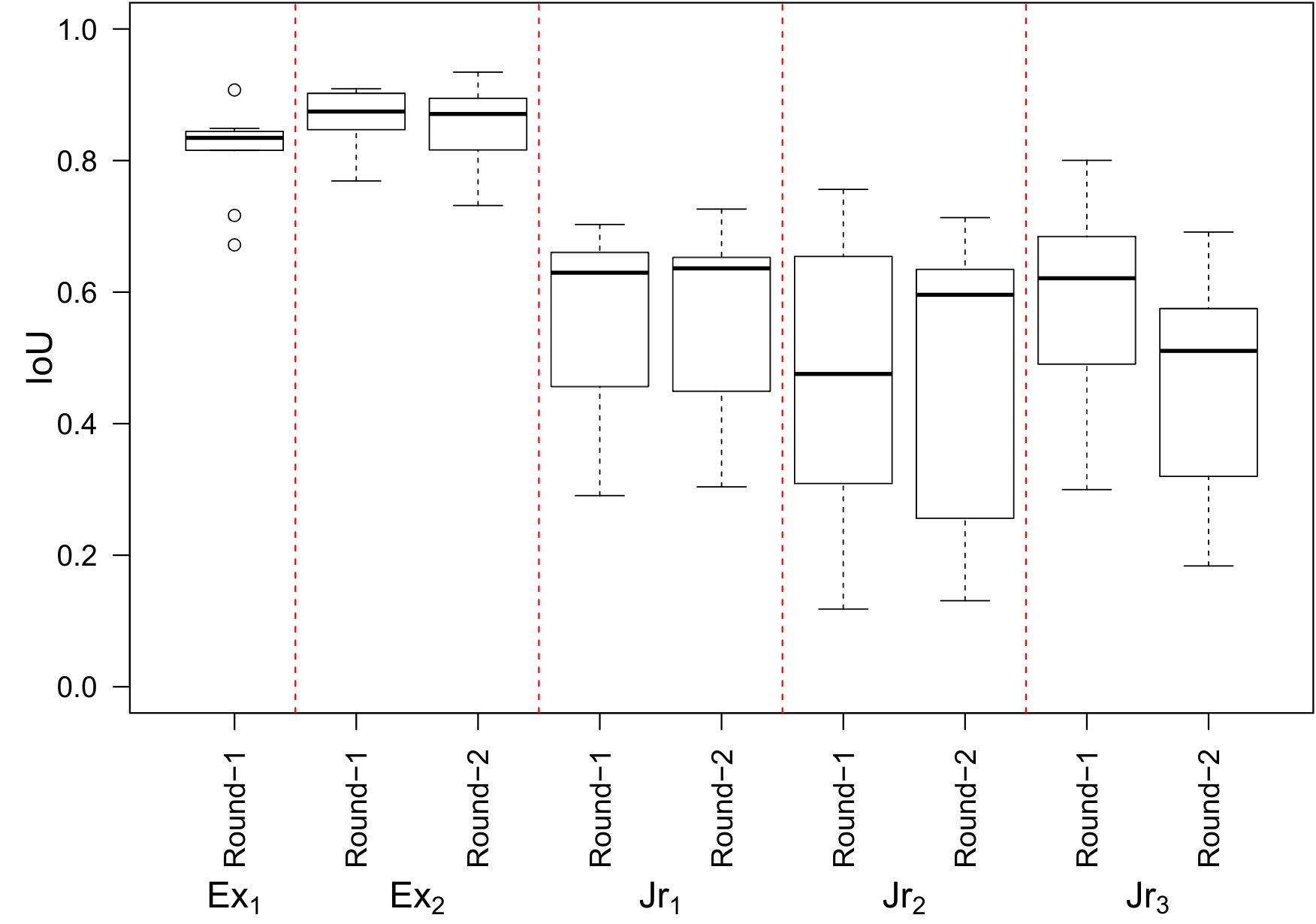
We evaluate the quality of diabetic macular edema (DME) intraretinal fluid (IRF) biomarker image annotations on OCT B-scans from five clinicians with a range of experience. Our investigation shows a notable variance in annotation performance, with a correlation that depends on the clinician's experience with OCT image interpretation of DME.
Image Processing
Multi-Modal Image Integration
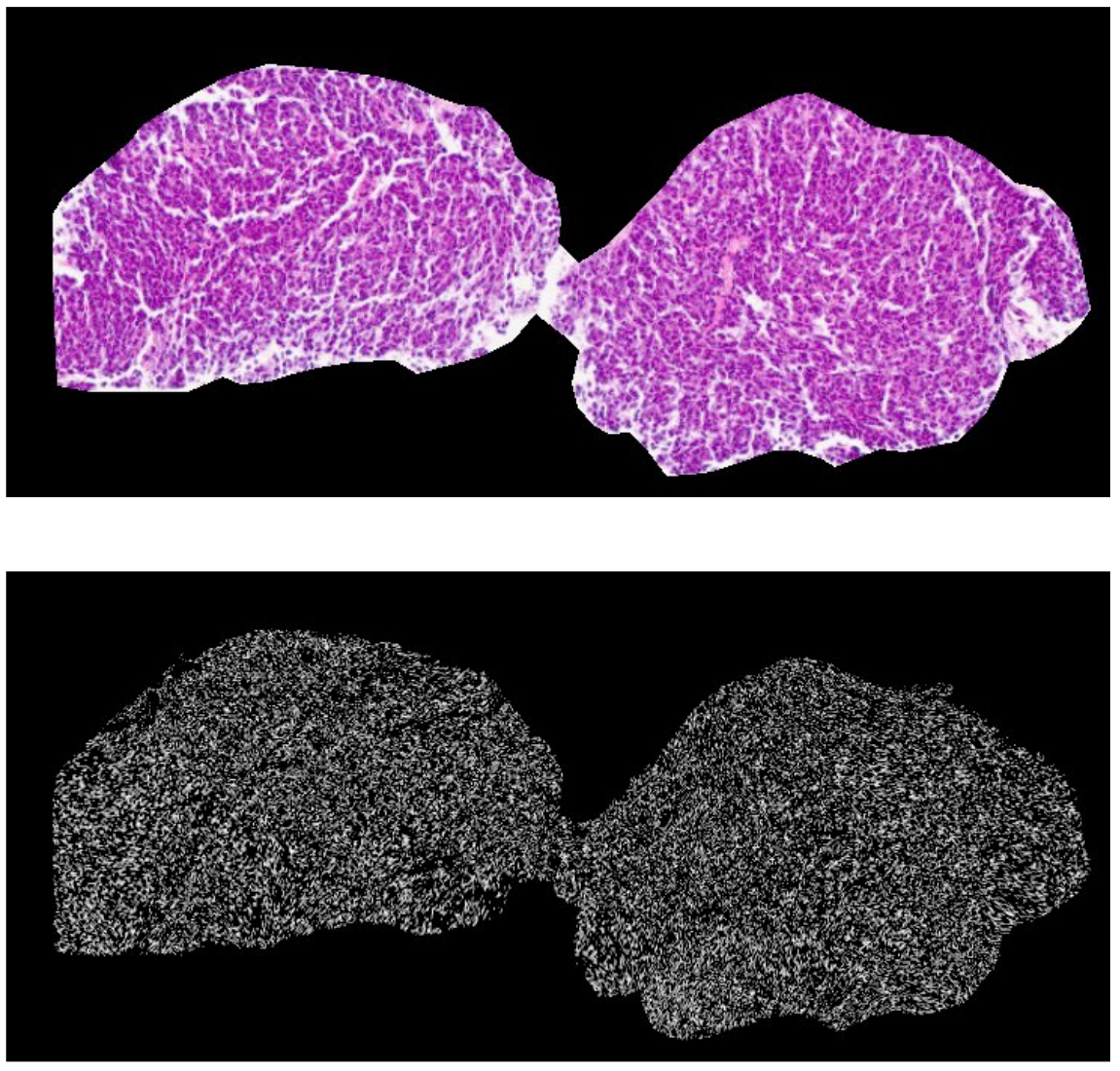
Our co-registration of Hematoxylin and Eosin (H&E), Laser Ablation-Inductively Coupled Plasma-Mass Spectrometry (LA-ICP-MS), and Imaging Mass Cytometry (IMC) images provides complementary information about the cellular and molecular characteristics of cancer tissue, enabling a more comprehensive understanding of the disease.
Image Processing
External Limiting Membrane
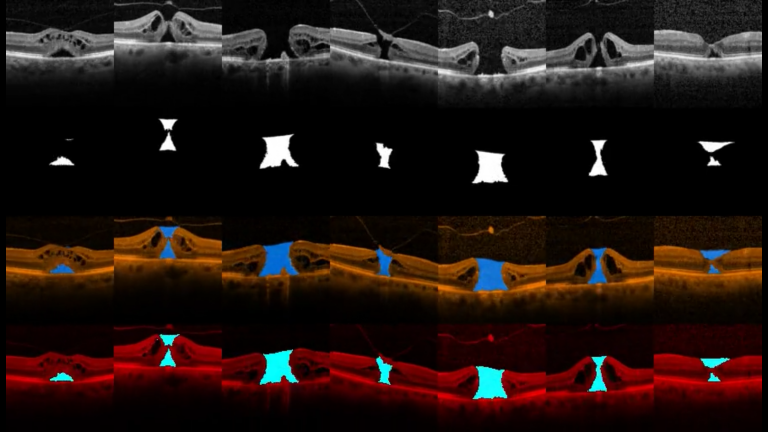
We design a new benchmark for the segmentation of the retinal external limiting membrane (ELM) using an image dataset of spectral domain optical coherence tomography (OCT) scans in a patient population with idiopathic full-thickness macular holes. Then, we compared qualitative and quantitative results with seven state-of-the-art machine learning-based segmentation methods to identify the ELM line with an automated system.
Image Processing
Macular Oedema
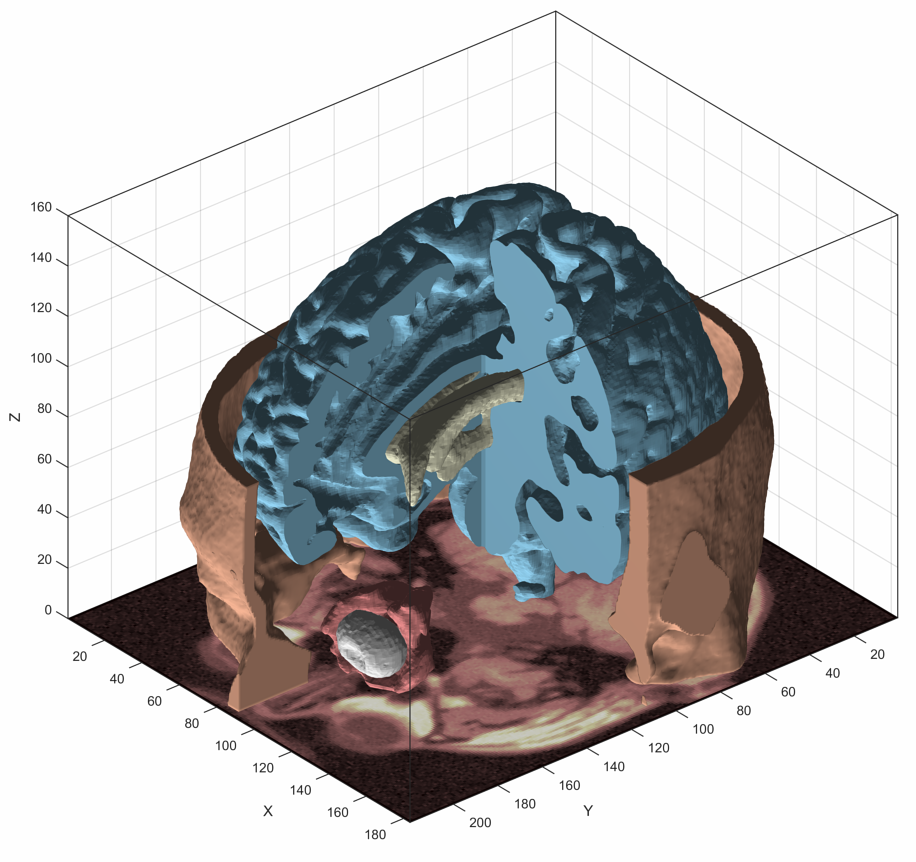
The objectives of this project are to develop and assess novel image analysis techniques of 3D OCT scans for volumetric assessment of different morphological patterns in diabetic macular oedema (DMO) such as: 1) Overall volume of the generalised outer retinal thickening in early DMO. 2) Volume of accumulated serous fluid under the neurosensory retina (SRF). 3) Residual volume of retinal tissue passing between the inner and outer plexiform retinal layers, which is an optimal measurement of potential residual macular function.
Image Processing
Fungal Networks
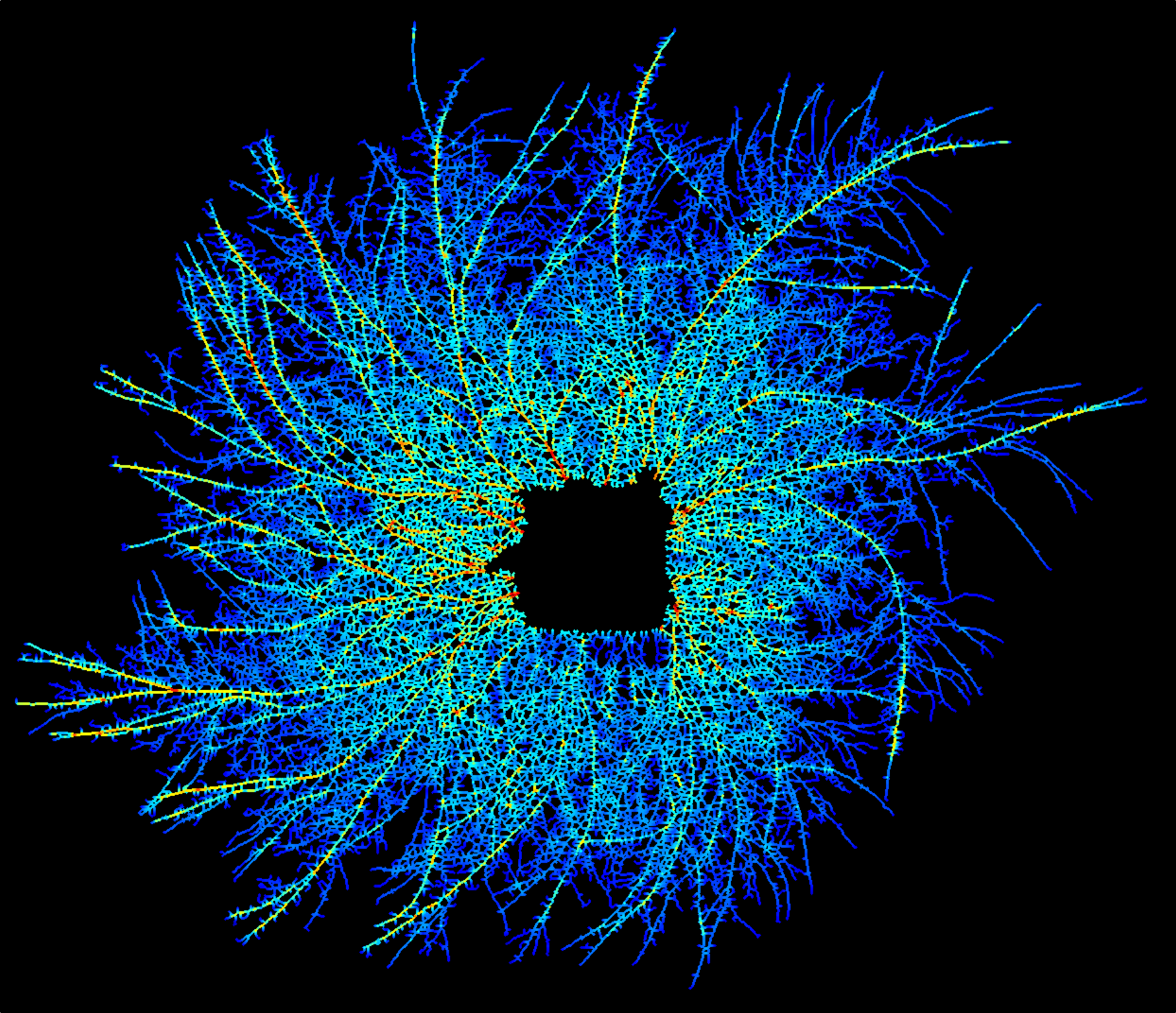
Saprotrophic fungi are critical in ecosystem biology as they are the only organisms capable of complete degradation of wood in temperate forests. The architecture of the network continuously adapts to local nutritional cues, damage or predation, through growth, branching, fusion or regression. We propose an image processing approach for fungal network segmentation and graph-based analysis.
Image Processing
Cytoskeletal Networks
A 3D/4D cytoskeletal network may contain thousands of nodes and connections, in specific but variable geometric organisations which vary with time. Such data exceeds the capacity of human analysis, creating a barrier, which can only be overcome with robust, automated image analysis and informatics tools to extract, characterise and model networks.
Image Processing
Graphs
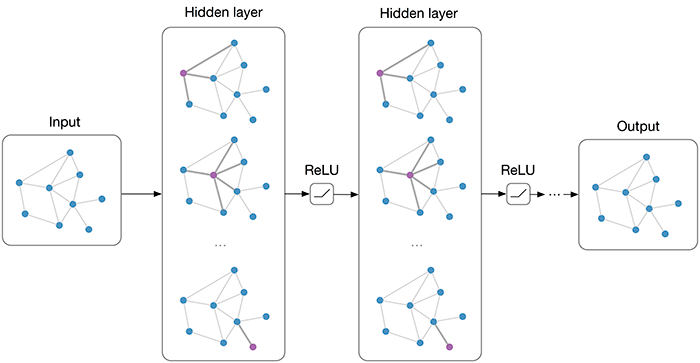
Graph embedding techniques aim to automatically create a low-dimensional representation of a given graph, which captures key structural elements in the resulting embedding space. We demonstrate that several topological features are indeed being approximated by the embedding space, allowing key insight into how graph embeddings create good representations.
Best Practices
GPU-Based Segmentation
The local Gaussian distribution fitting energy model is a state-of-the-art method, capable of segmenting inhomogeneous objects with poorly defined boundaries, but it is computationally expensive. In our approach, we port the energy functional to the GPU, and introduce a novel set of interactive brush functions to segment challenging datasets where an active contour would not ordinarily capture the target object.
Image Processing
Helix
This project focuses on a robust method for extracting a natural interpolating 3D piecewise cubic spline from a 2D input image of a helix object. We are able to use the generated spline to extract 3D metrics such as tortuosity and curvature. The generated 3D spline has few input parameters, and only requires a single view of the 2D dataset, making it suitable for a range of applications in engineering, physics, and biology.
Best Practices
Big Data
Projects focuses on big data challenges including capturing data, data storage, data analysis, search, sharing, transfer, visualization, querying, updating and information privacy.
Image Processing
Collocalisation
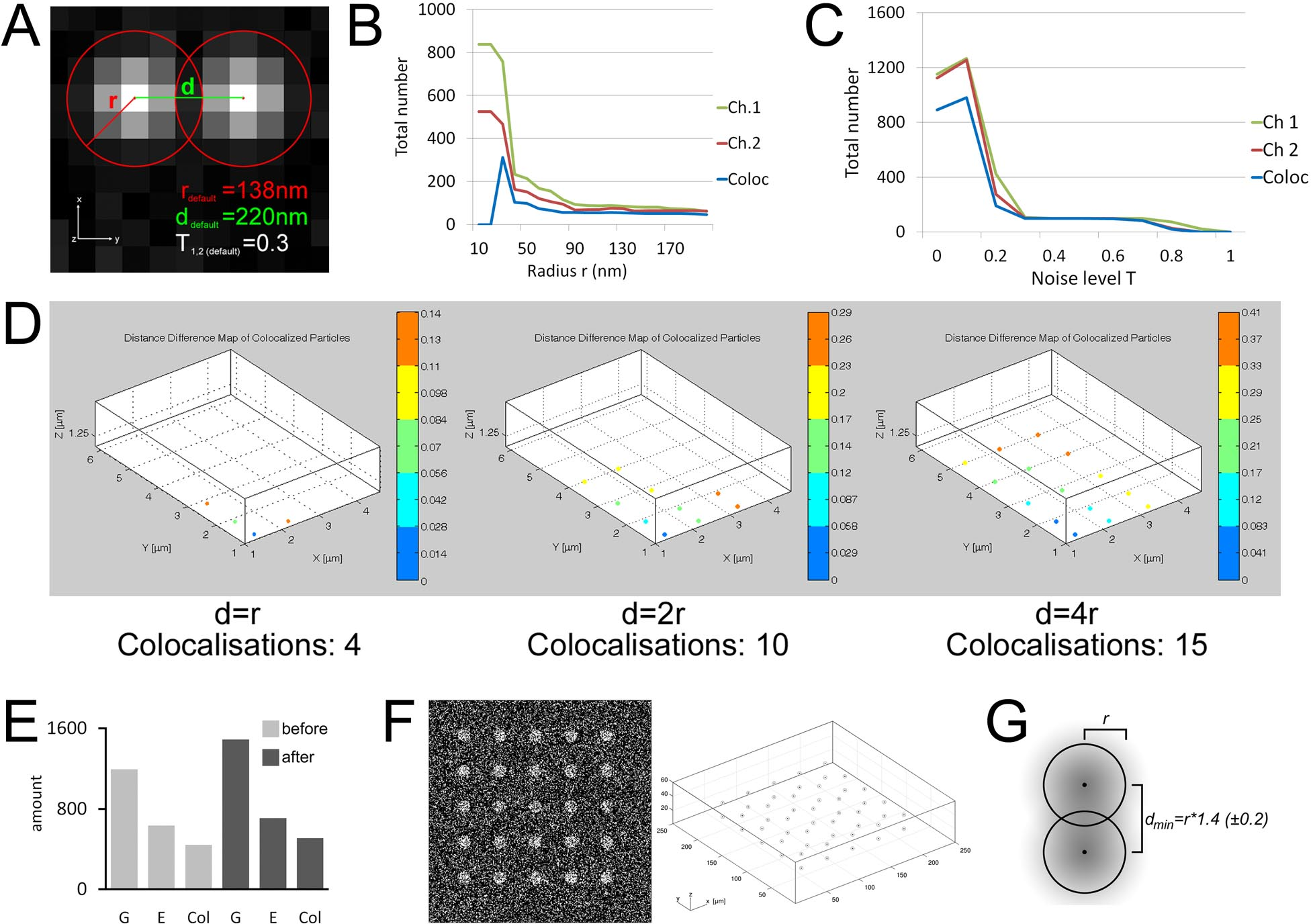
The colocalization technique is important to many cell biological and physiological studies during the demonstration of a relationship between pairs of bio-molecules.
Image Processing
Deep Learning
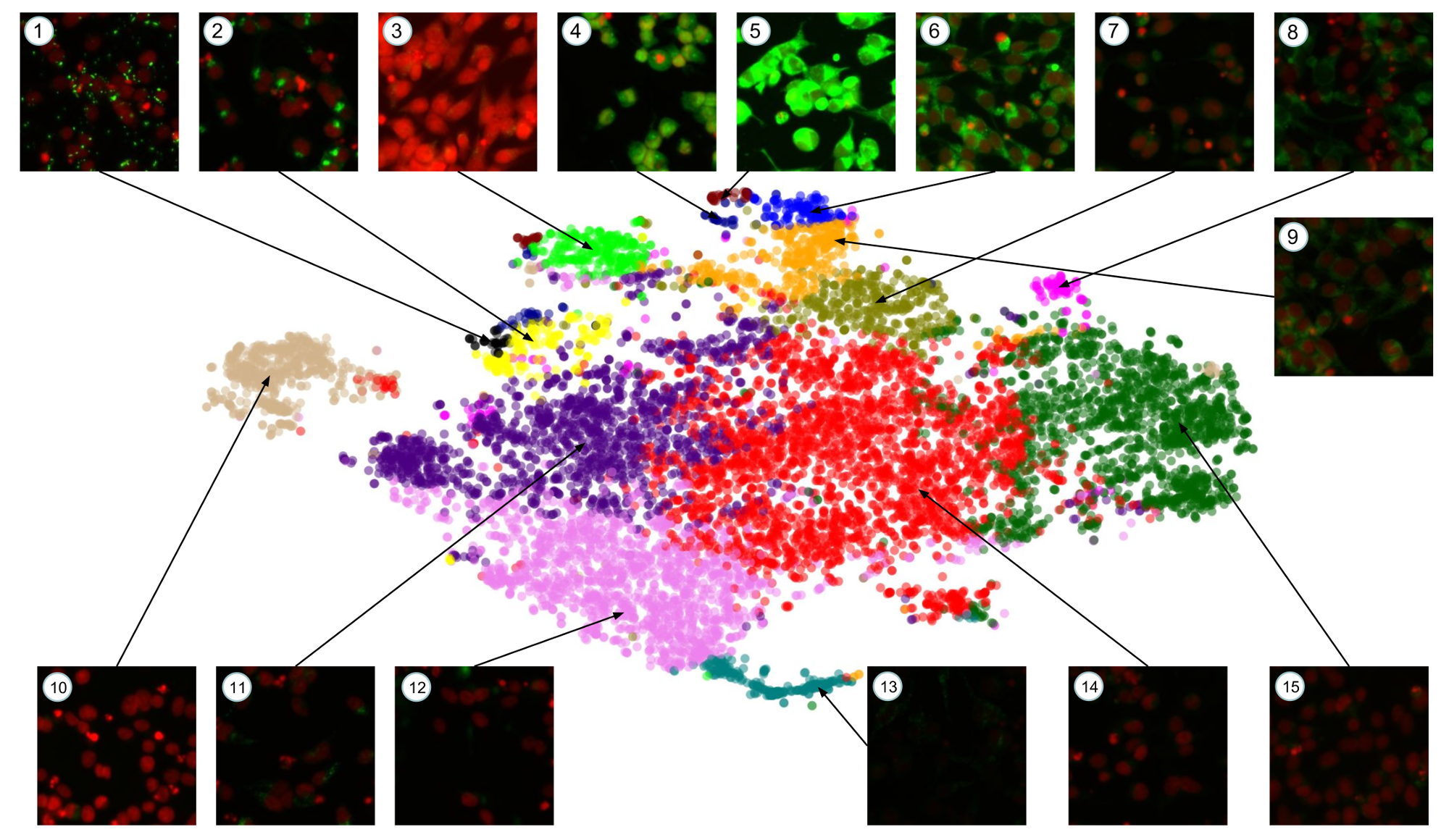
In this paper, we show how pre-trained convolutional image features can be used to assist scientists in discovering interesting chemical clusters for further investigation. Our method reduces the dimensionality of raw fluorescent stained images from a high throughput imaging (HTI) screen, producing an embedding space that groups together images with similar cellular phenotypes. Running standard unsupervised clustering on this embedding space yields a set of distinct phenotypic clusters. This allows scientists to further select and focus on interesting clusters for downstream analyses.
Image Processing
Ellipse
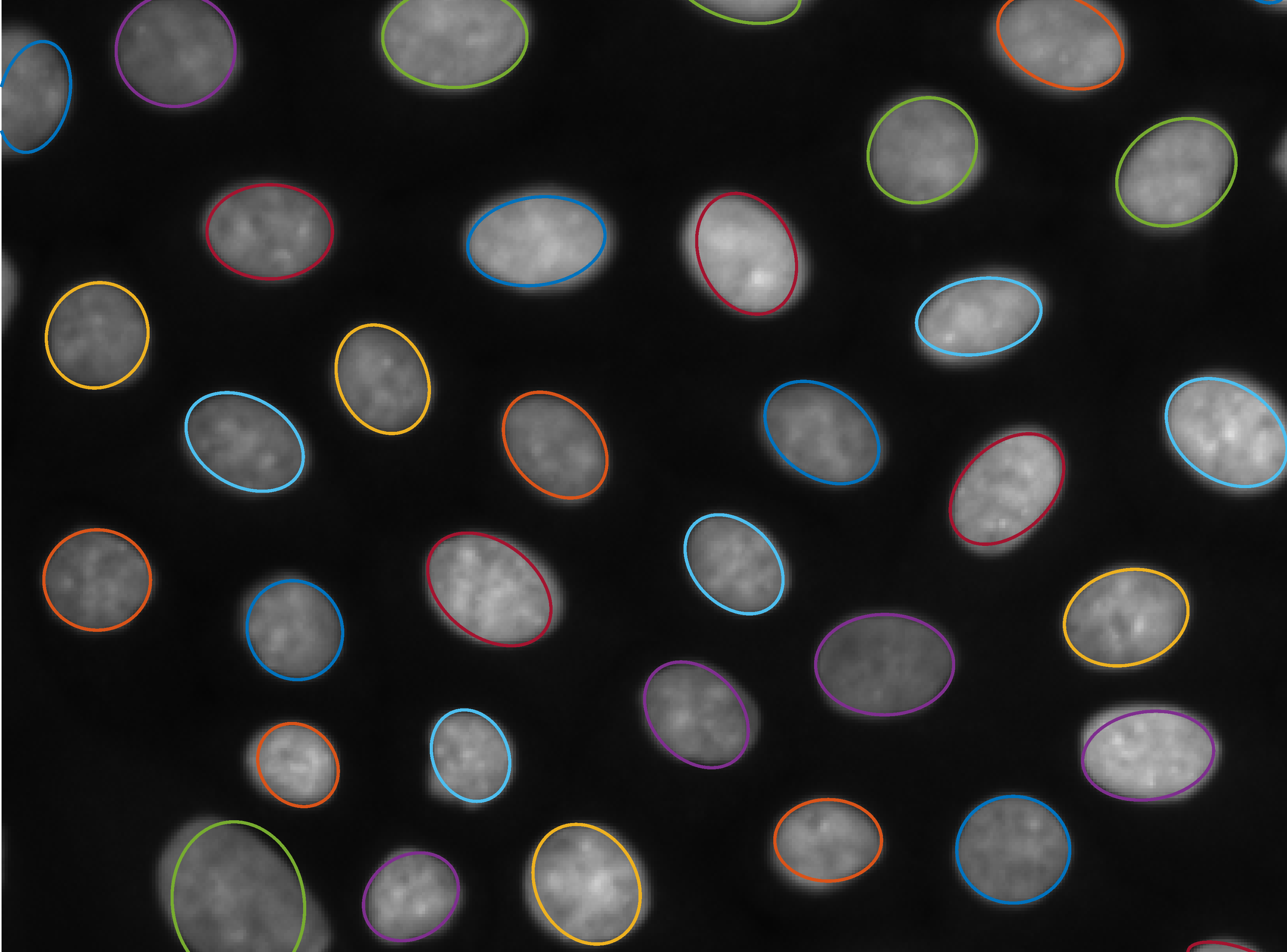
We introduce a new method for the detection of parametrisable shapes in N dimensions; this method has been developed for ellipses and uses simple morphological operations, building on aspects of granulometry and signal processing techniques. With this method it is possible to easily extract the position, size and rotation of elliptical objects in any image data. This method is low parameter, accurate and robust to noise and object clustering in greyscale images; key contributions are the abiltiy to find an unknown number of ellipses with no a priori information and no arbitrary thresholding and robustness to both noise and clustering.
Image Processing
Nanotubes
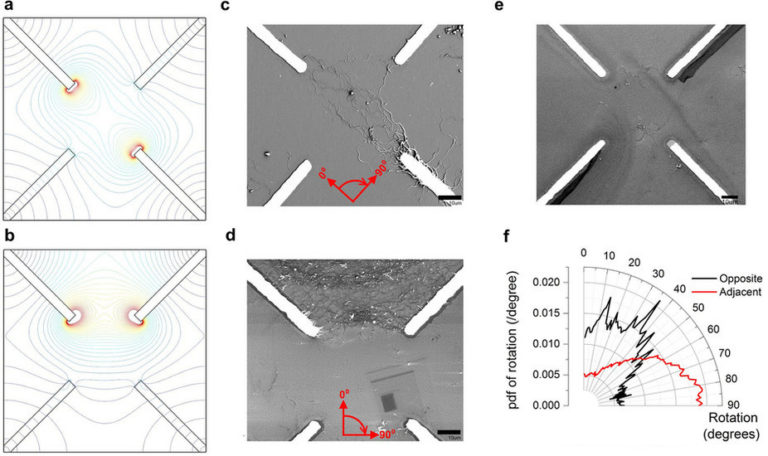
In this project we demonstrate the evolution of a disordered composite material, using voltages as the external stimuli, into a form where a simple computational problem can be solved. The material consists of single-walled carbon nanotubes suspended in liquid crystal; the nanotubes act as a conductive network, with the liquid crystal providing a host medium to allow the conductive network to reorganise when voltages are applied. We show that the application of electric fields under computer control results in a significant change in the material morphology, favouring the solution to a classification task.
Image Processing
Bacteria
Differential interference contrast microscopy plays an important role in modern bacterial cell biology. We have developed and evaluated a high-throughput image analysis and processing approach to detect and characterize bacterial cells and chemotaxis proteins. Its performance was evaluated using differential interference contrast and fluorescence microscopy images of Rhodobacter sphaeroides.
Image Processing
Cell Tip Tracking
Fungi cause devastating plant and human diseases. There is considerable evidence that much of the cellular machinery driving growth of invasive fungal hyphae is common across all fungi, including plant and mammalian pathogens, and involves localized tip growth. We have developed and evaluated high-throughput automated microscope-based multi-dimensional image analysis systems to segment and characterize fungal growth, and characterize the patterns of protein localization within the tip that control development.
Image Processing
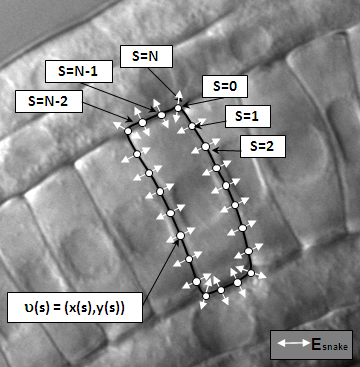
Ascidian Cells Tracking
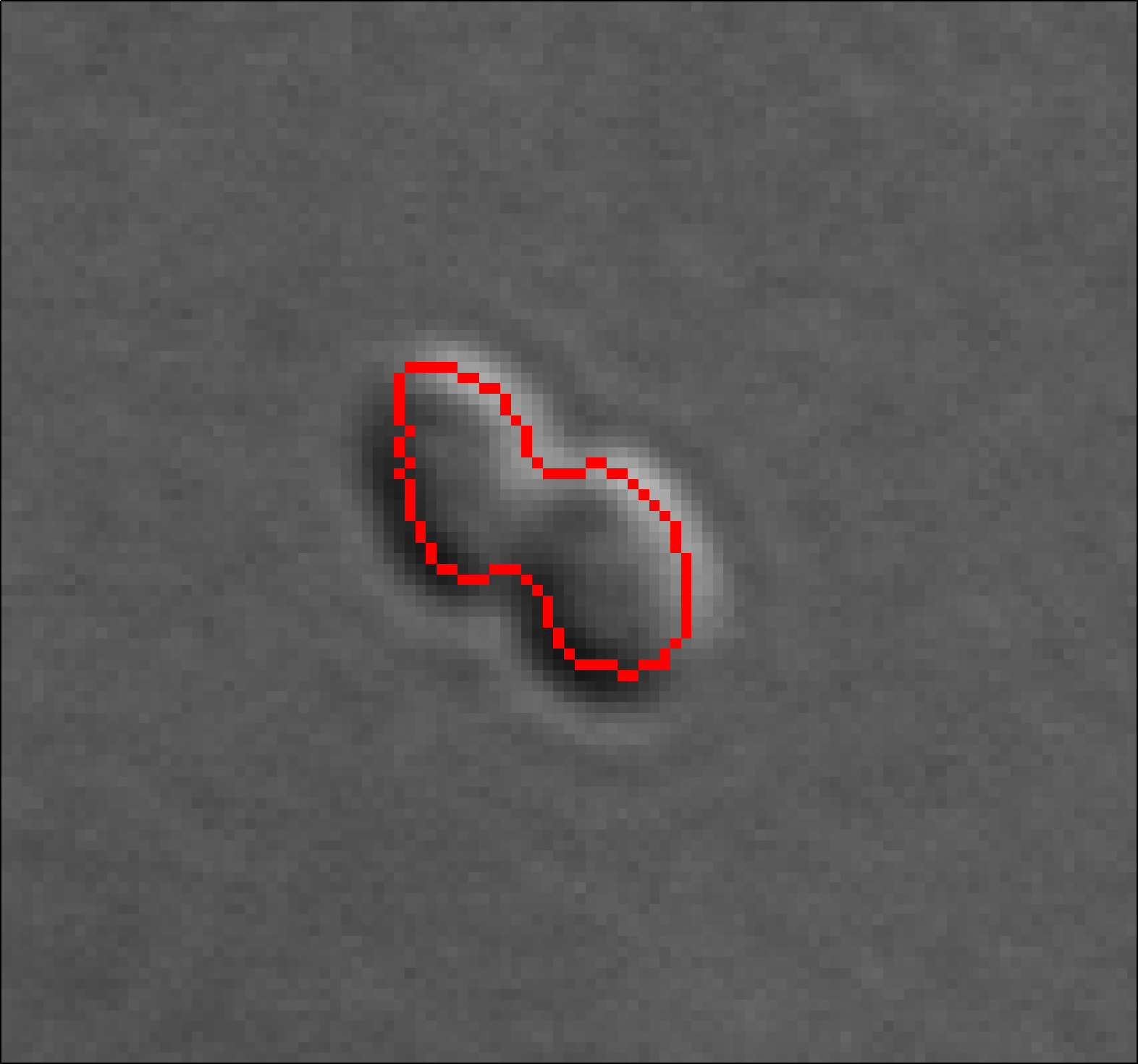
A method to automatically segment notochord cell boundaries from differential interference contrast (DIC) timelapse images of the elongating ascidian tail. The method is based on a specialized parametric active contour, the network snake, which can be initialized as a network of arbitrary but fixed topology and provides an effective framework for simultaneously segmenting multiple touching cells.
Image Processing
Cancer Cell Protrusions
Identification, description and quantification of the cell behavior observed by time-lapse confocal microscopy Research, development and implementation of novel 4D image analysis and processing methods for identification, description and quantification of the cell behavior (protrusion, adhesion and invasion), as observed by time-lapse confocal microscopy.
Image Processing
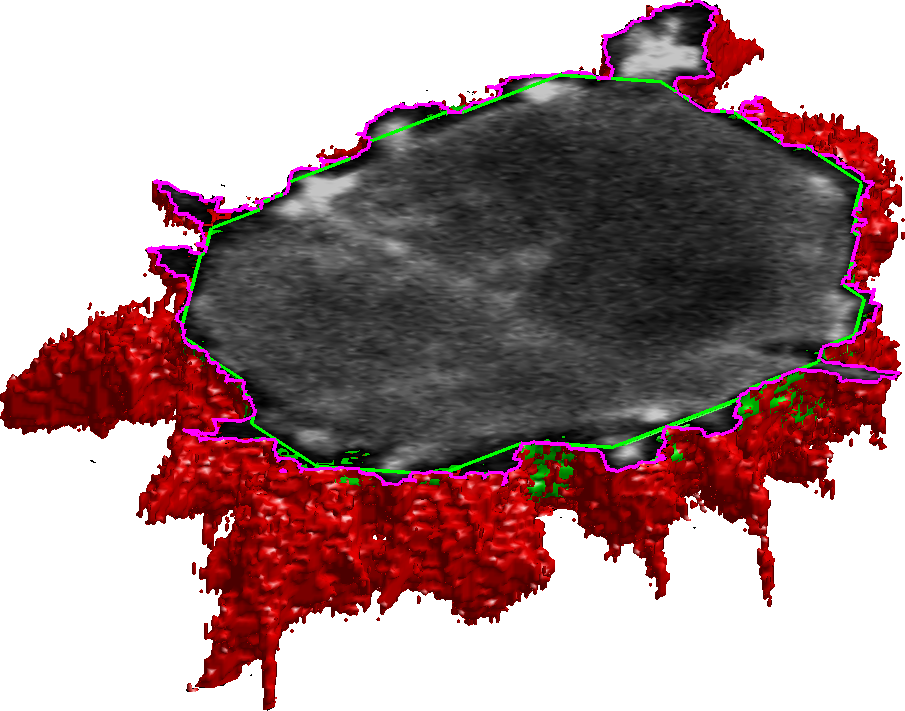
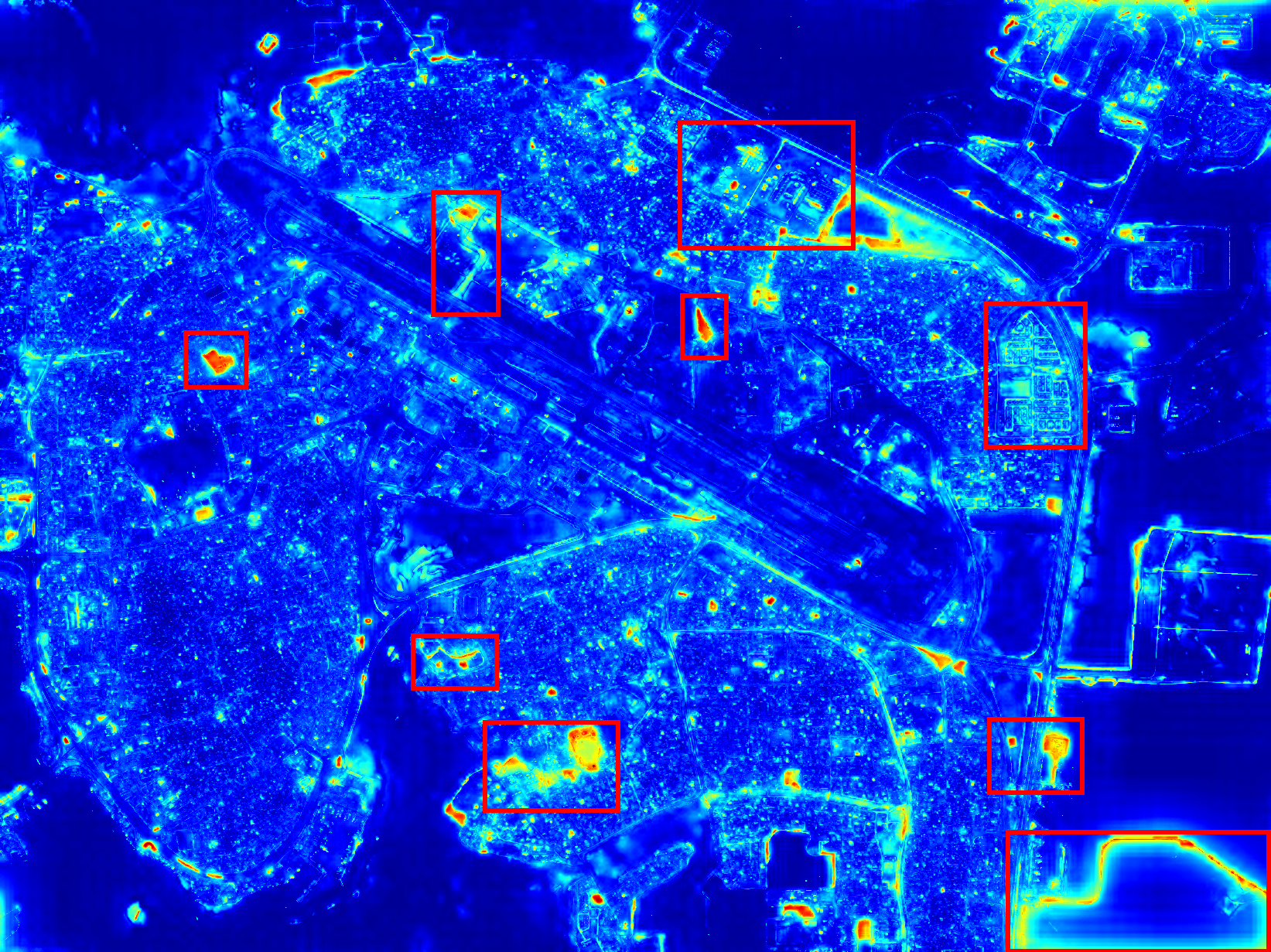
Change Detection
Runways are vital descriptive features of airports and knowledge of their location is important to many aviation and military applications. With the recent wide availability of remote sensing data, there is demand for an automatic process of extracting runway geometry from satellite imagery. In this project we establish a novel method for accurate and precise extraction of geometric polygons for an arbitrary number of runways in VHR remote sensing imagery.
Best Practices
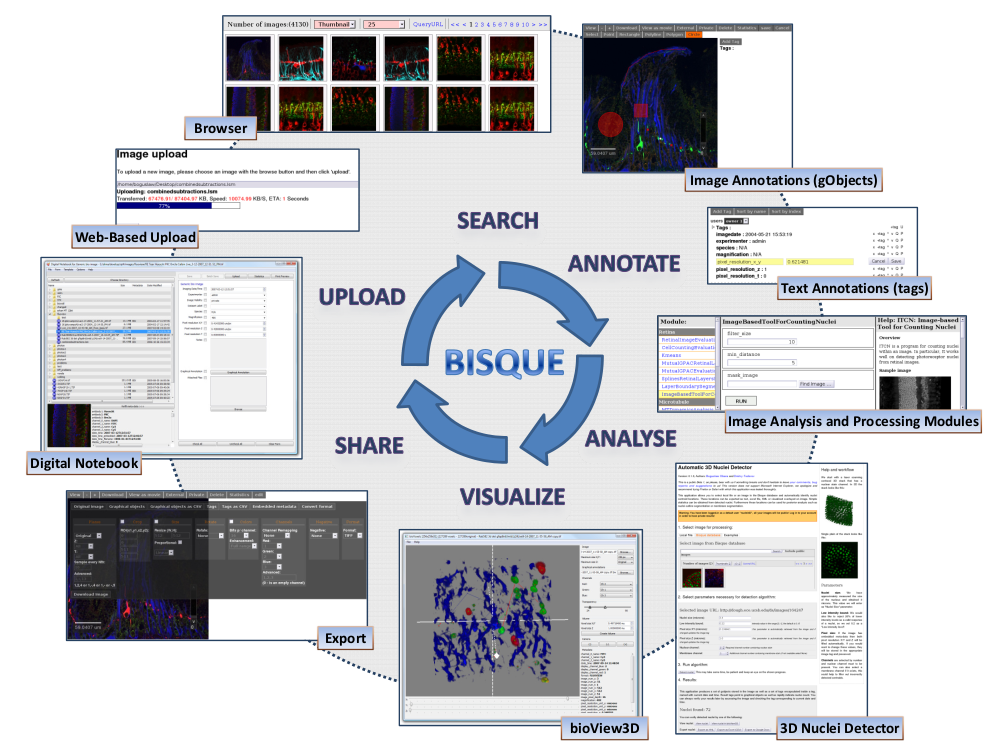
Bisque
Bisque provides a platform for data sharing, flexible metadata management, and integrated extensible image analysis and processing. Bisque system is used by many research and commercial institutions including: 1) iPlant: a community of researchers, educators, and students working to enrich all plant sciences through the development of cyberinfrastructure that are essential components of modern biology. 2) Pfizer - The Pfizer Neuroscience Research Unit is developing quantitative methods that process the images, keeping track of the processing workflow, and allowing scientists to create new work flows that seamlessly combine multimodal data.